[ad_1]
A finalist for the Gordon Bell particular prize for prime efficiency computing-based COVID-19 analysis has taught giant language fashions (LLMs) a brand new lingo — gene sequences — that may unlock insights in genomics, epidemiology and protein engineering.
Printed in October, the groundbreaking work is a collaboration by greater than two dozen educational and industrial researchers from Argonne Nationwide Laboratory, NVIDIA, the College of Chicago and others.
The analysis staff educated an LLM to trace genetic mutations and predict variants of concern in SARS-CoV-2, the virus behind COVID-19. Whereas most LLMs utilized to biology so far have been educated on datasets of small molecules or proteins, this venture is without doubt one of the first fashions educated on uncooked nucleotide sequences — the smallest items of DNA and RNA.
“We hypothesized that shifting from protein-level to gene-level information may assist us construct higher fashions to know COVID variants,” mentioned Arvind Ramanathan, computational biologist at Argonne, who led the venture. “By coaching our mannequin to trace your complete genome and all of the modifications that seem in its evolution, we will make higher predictions about not simply COVID, however any illness with sufficient genomic information.”
The Gordon Bell awards, thought to be the Nobel Prize of excessive efficiency computing, shall be offered at this week’s SC22 convention by the Affiliation for Computing Equipment, which represents round 100,000 computing consultants worldwide. Since 2020, the group has awarded a particular prize for excellent analysis that advances the understanding of COVID with HPC.
Coaching LLMs on a 4-Letter Language
LLMs have lengthy been educated on human languages, which often comprise a pair dozen letters that may be organized into tens of hundreds of phrases, and joined collectively into longer sentences and paragraphs. The language of biology, alternatively, has solely 4 letters representing nucleotides — A, T, G and C in DNA, or A, U, G and C in RNA — organized into completely different sequences as genes.
Whereas fewer letters could seem to be a less complicated problem for AI, language fashions for biology are literally much more sophisticated. That’s as a result of the genome — made up of over 3 billion nucleotides in people, and about 30,000 nucleotides in coronaviruses — is tough to interrupt down into distinct, significant items.
“On the subject of understanding the code of life, a significant problem is that the sequencing info within the genome is kind of huge,” Ramanathan mentioned. “The which means of a nucleotide sequence may be affected by one other sequence that’s a lot additional away than the following sentence or paragraph can be in human textual content. It may attain over the equal of chapters in a e-book.”
NVIDIA collaborators on the venture designed a hierarchical diffusion methodology that enabled the LLM to deal with lengthy strings of round 1,500 nucleotides as in the event that they had been sentences.
“Commonplace language fashions have hassle producing coherent lengthy sequences and studying the underlying distribution of various variants,” mentioned paper co-author Anima Anandkumar, senior director of AI analysis at NVIDIA and Bren professor within the computing + mathematical sciences division at Caltech. “We developed a diffusion mannequin that operates at a better stage of element that permits us to generate practical variants and seize higher statistics.”
Predicting COVID Variants of Concern
Utilizing open-source information from the Bacterial and Viral Bioinformatics Useful resource Middle, the staff first pretrained its LLM on greater than 110 million gene sequences from prokaryotes, that are single-celled organisms like micro organism. It then fine-tuned the mannequin utilizing 1.5 million high-quality genome sequences for the COVID virus.
By pretraining on a broader dataset, the researchers additionally ensured their mannequin may generalize to different prediction duties in future tasks — making it one of many first whole-genome-scale fashions with this functionality.
As soon as fine-tuned on COVID information, the LLM was in a position to distinguish between genome sequences of the virus’ variants. It was additionally in a position to generate its personal nucleotide sequences, predicting potential mutations of the COVID genome that would assist scientists anticipate future variants of concern.
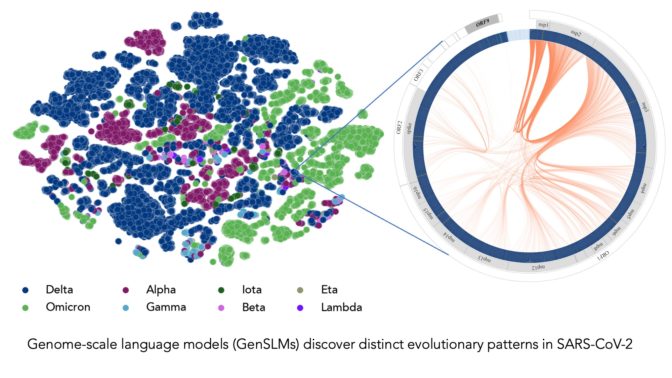
“Most researchers have been monitoring mutations within the spike protein of the COVID virus, particularly the area that binds with human cells,” Ramanathan mentioned. “However there are different proteins within the viral genome that undergo frequent mutations and are essential to know.”
The mannequin may additionally combine with fashionable protein-structure-prediction fashions like AlphaFold and OpenFold, the paper acknowledged, serving to researchers simulate viral construction and examine how genetic mutations affect a virus’ capacity to contaminate its host. OpenFold is without doubt one of the pretrained language fashions included within the NVIDIA BioNeMo LLM service for builders making use of LLMs to digital biology and chemistry functions.
Supercharging AI Coaching With GPU-Accelerated Supercomputers
The staff developed its AI fashions on supercomputers powered by NVIDIA A100 Tensor Core GPUs — together with Argonne’s Polaris, the U.S. Division of Power’s Perlmutter, and NVIDIA’s in-house Selene system. By scaling as much as these highly effective techniques, they achieved efficiency of greater than 1,500 exaflops in coaching runs, creating the biggest organic language fashions so far.
“We’re working with fashions at present which have as much as 25 billion parameters, and we anticipate this to considerably improve sooner or later,” mentioned Ramanathan. “The mannequin dimension, the genetic sequence lengths and the quantity of coaching information wanted means we actually want the computational complexity supplied by supercomputers with hundreds of GPUs.”
The researchers estimate that coaching a model of their mannequin with 2.5 billion parameters took over a month on round 4,000 GPUs. The staff, which was already investigating LLMs for biology, spent about 4 months on the venture earlier than publicly releasing the paper and code. The GitHub web page contains directions for different researchers to run the mannequin on Polaris and Perlmutter.
The NVIDIA BioNeMo framework, obtainable in early entry on the NVIDIA NGC hub for GPU-optimized software program, helps researchers scaling giant biomolecular language fashions throughout a number of GPUs. A part of the NVIDIA Clara Discovery assortment of drug discovery instruments, the framework will help chemistry, protein, DNA and RNA information codecs.
Discover NVIDIA at SC22 and watch a replay of the particular tackle beneath:
Picture at high represents COVID strains sequenced by the researchers’ LLM. Every dot is color-coded by COVID variant. Picture courtesy of Argonne Nationwide Laboratory’s Bharat Kale, Max Zvyagin and Michael E. Papka.
[ad_2]
